Center for Personalized Diagnostics
Penn Medicine's Center for Personalized Diagnostics (CPD) utilizes massively parallel sequencing panels to simultaneously evaluate multiple targetable genes in cancer specimens. Supporting the clinical oncology team at Penn, our panels provide information to identify individualized therapies that can dramatically alter a patient's treatment course and outcome.
CPD currently offers the following gene panels (click tables for larger view):
PennSeqTM Hematologic Malignancies Panel
Sequence analysis of 202 genes

- Accepted Specimens: Blood; bone marrow; formalin-fixed, paraffin-embedded (FFPE) tissue; fresh tissue in PreservCyt
- Minimum Requirements: 10% tumor nuclei for tissue; 100ng DNA (non-FFPE), 200ng DNA (FFPE)
- Covers: Genes listed for the entire coding sequence +/- ~8bp flanking intronic sequence; two hotspots in the TERT promoter; Select Hotspot variants only in U2AF1
- Detects: Single nucleotide variants (SNVs); small indels; copy number gains in ABL1, PDGFRA, and MYC
- Limitations: Lower limit of reportability 4% variant allele fraction (VAF) [1% for FLT3 ITDs only]. No deep intronic splice variants; no promoter variants outside of TERT; no structural rearrangements; no methylation; no copy number loss
The PennSeq assay enriches for many genomic regions and then is computationally filtered to report variants from a curated set of genes. Incidental variants may be discovered during routine review of the genomic data that fall outside of the curated set of genes. Any Disease-Associated or other potentially significant variant discovered incidentally will be included in the clinical report with a comment and appropriate interpretive text, as needed. By requesting this testing and accepting this report for clinical purpose, implied consent is given that information on incidentally identified genomic variants may be received.
Hematological Expedited Sequencing (HEXS) Panel
DNA sequencing analysis of single nucleotide variants (SNVs) and insertions/deletions (indels) in 58 genes [18 full coding sequence (CDS) genes; 40 hotspot genes].

- Accepted Specimens: Blood; bone marrow; bone marrow core in saline
- Minimum Requirements: 20 ng genomic DNA
- Covers: DNA full CDS and hotspots as indicated in the table above
- Detects: SNVs, indels, and FLT3-Internal Tandem Duplications (FLT3-ITDs)
- Limitations: Lower limit of reportability 2% variant allele fractions (VAFs) for SNVs, indels <10bp, and FLT3-ITDS <21bp; 1% VAF for indels ≥10bp; and 0.3% VAF for FLT3-ITDs ≥21bp up to 84bp. FLT3-ITDs > 84bp may not be detected. No structural rearrangements; no methylation; no copy number changes; no tumor mutational burden.
PennSeqTM Solid Tumor Panel
Sequence analysis of 211 genes
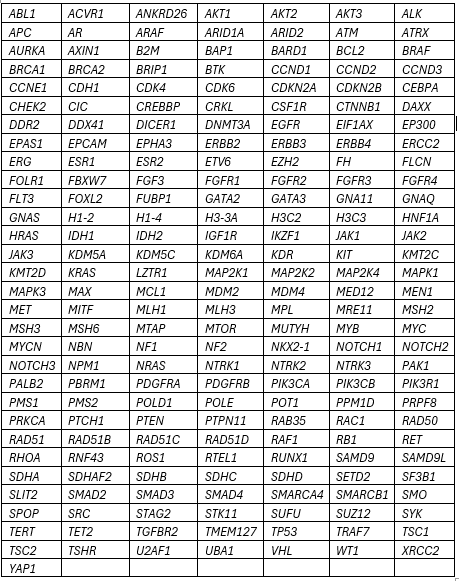
- Accepted Specimens: Blood; bone marrow; formalin-fixed, paraffin-embedded (FFPE) tissue; fresh tissue in PreservCyt
- Minimum Requirements: 10% tumor nuclei for tissue; 100ng DNA (non-FFPE), 200ng DNA (FFPE)
- Covers: Genes listed for the entire coding sequence +/- ~8bp flanking intronic sequence; two hotspots in the TERT promoter; Select Hotspot variants only in U2AF1
- Detects: Single nucleotide variants (SNVs); small indels; Targeted copy number gains
- Limitations: Lower limit of reportability 4% variant allele fraction (VAF) [1% for FLT3 ITDs only]. No deep intronic splice variants; no promoter variants outside of TERT; no structural rearrangements; no methylation; no copy number loss
The PennSeq assay enriches for many genomic regions and then is computationally filtered to report variants from a curated set of genes. Incidental variants may be discovered during routine review of the genomic data that fall outside of the curated set of genes. Any Disease-Associated or other potentially significant variant discovered incidentally will be included in the clinical report with a comment and appropriate interpretive text, as needed. By requesting this testing and accepting this report for clinical purpose, implied consent is given that information on incidentally identified genomic variants may be received.
Fusion Transcript Panel
RNA sequencing analysis of 56 genes for rearrangements
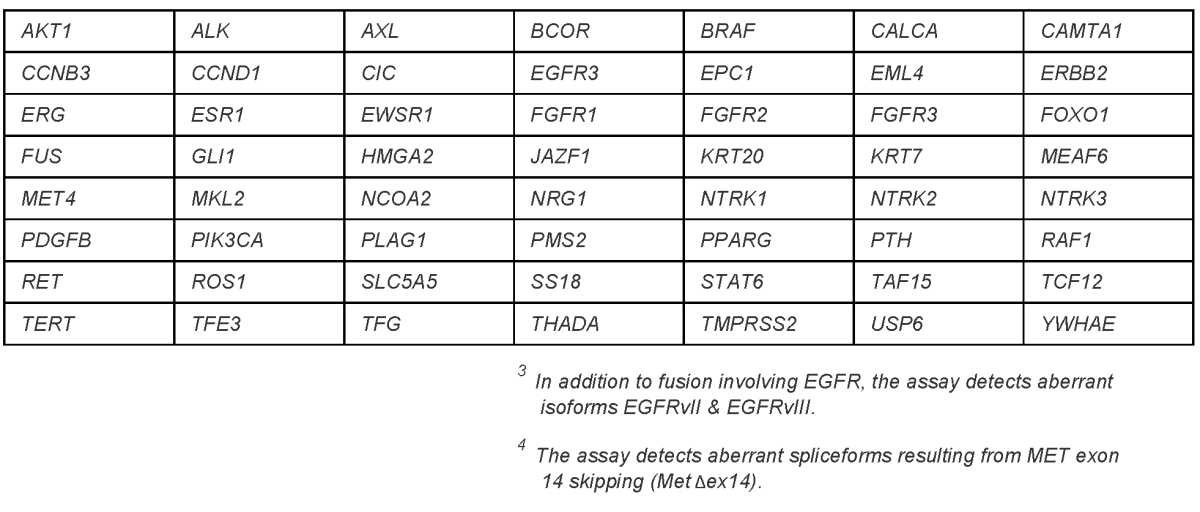
- Accepted Specimens: Formalin-fixed, paraffin-embedded (FFPE) tissue; fresh tissue in PreservCyt
- Minimum Requirements: 10% neoplastic tissue
- Covers: Selected exon-intron boundaries
- Detects: Aberrant transcripts involving the included exons; can detect novel fusion partners at known break-points
- Limitations: Only detects fusions which include at least one of the targets at the included exons; no SNVs; no copy number changes; no small indels; no methylation
Penn Precision Panel 2.0
Sequence analysis of 59 genes
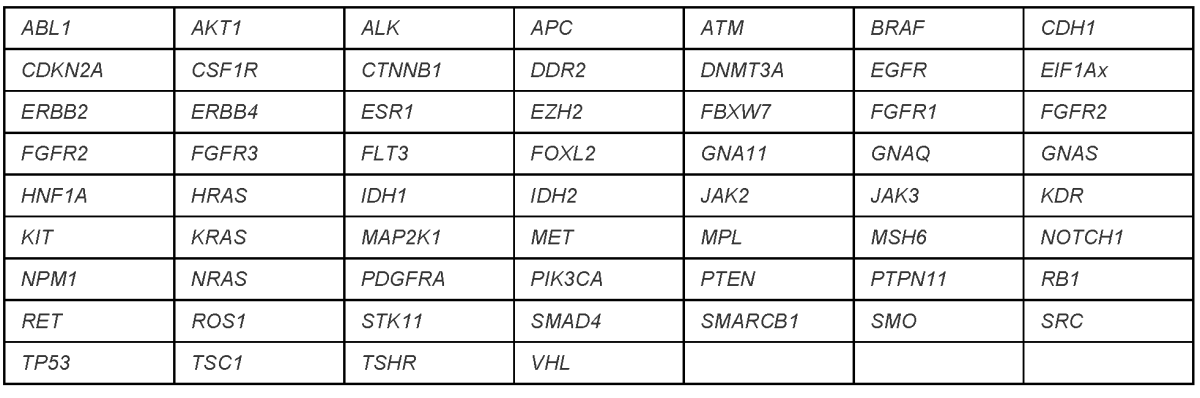
- Accepted Specimens: Formalin-fixed, paraffin-embedded (FFPE) tissue; fresh tissue in PreservCyt; blood; bone marrow
- Minimum Requirements: 10% neoplastic tissue; 100ng DNA (non-FFPE), 200ng DNA (FFPE)
- Covers: Hotspots and the entire coding sequence of TP53
- Detects: Single nucleotide variants (SNVs); small indels
- Limitations: Lower limit of reportability 4% variant allele fraction; indels unreliable >20bp; no deep intronic splice variants; no structural rearrangements; no copy number variants; no methylation
Targeted Expedited Molecular Profiling (TEMP)
DNA sequence analysis of single nucleotide variant (SNV) and insertion/deletion (indel) hotspots in 45 genes. RNA sequence analysis of 13 fusion drivers and 2 oncogenic isoforms.

- Accepted Specimens: Formalin-fixed paraffin-embedded (FFPE) tissue, cytology samples, FNA rinses, pleural effusion/fluid, and fresh tissue
- Minimum Requirements: 10% neoplastic tissue
- Covers: DNA hotspots and selected fusions and oncogenic isoforms in the genes listed above
- Detects: SNVs, small indels, targeted fusions and oncogenic isoforms with known breakpoints and non-targeted fusions with unknown breakpoints
- Limitations: Only detects known hotspot SNV and indel variants and known fusions/oncogenic isoforms; lower limit of reportability of 2.5% variant allele fraction (VAF) for SNVs, 2.0% VAF for indels; no methylation; no copy number changes



